stan_glm
family: gaussian [identity]
formula: y ~ x
observations: 15
predictors: 2
------
Median MAD_SD
(Intercept) 0.1 1.4
x 2.1 0.2
Auxiliary parameter(s):
Median MAD_SD
sigma 2.6 0.5
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanregSMaC: Statistics, Math, and Computing
APSTA-GE 2006: Applied Statistics for Social Science Research
Eric.Novik@nyu.edu | Summer 2025 | Session 8
Session 8 Outline
- Statistical analysis workflow
- Simulating data for simple regression
- Binary predictor in a regression
- Adding continuous predictor
- Combining binary and continuous predictors
- Interpreting coefficients
- Adding interactions
- Uncertainty and predictions
- Assumptions of the regression analysis
- Regression modeling advice
\[ \DeclareMathOperator{\E}{\mathbb{E}} \DeclareMathOperator{\P}{\mathbb{P}} \DeclareMathOperator{\V}{\mathbb{V}} \DeclareMathOperator{\L}{\mathscr{L}} \DeclareMathOperator{\I}{\text{I}} \]
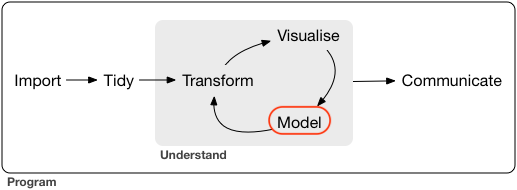
Analysis Workflow
- Following is a high-level, end-to-end view from “R for Data Science” by Wickham and Grolemund
Some Notation
- Observed data \(y\)
- Unobserved but observable data \(\widetilde{y}\)
- Unobservable parameters \(\theta\)
- Covariates \(X\)
- Prior distribution \(f(\theta)\) (for Bayesian inference)
- Data model or sampling distribution (as a function of y) \(f(y \mid \theta, X)\)
- Likelihood (as a function of \(\theta\)), \(f(y \mid \theta, X)\), sometimes written as \(\mathcal{L}(\theta \mid y, X)\)
- Maximum likelihood estimate (MLE): \(\hat{\theta} = \underset{\theta}{\text{argmax}} \, \mathcal{L}(\theta \mid y, X)\) (for Frequentist inference)
- Posterior distribution \(f(\theta \mid y, X)\) (for Bayesian inference)
Bayes vs Frequentist Inference
- Estimation is the process of figuring out the unknowns, i.e., unobserved quantities
- In frequentist inference, the problem is framed in terms of the most likely value(s) of \(\theta\)
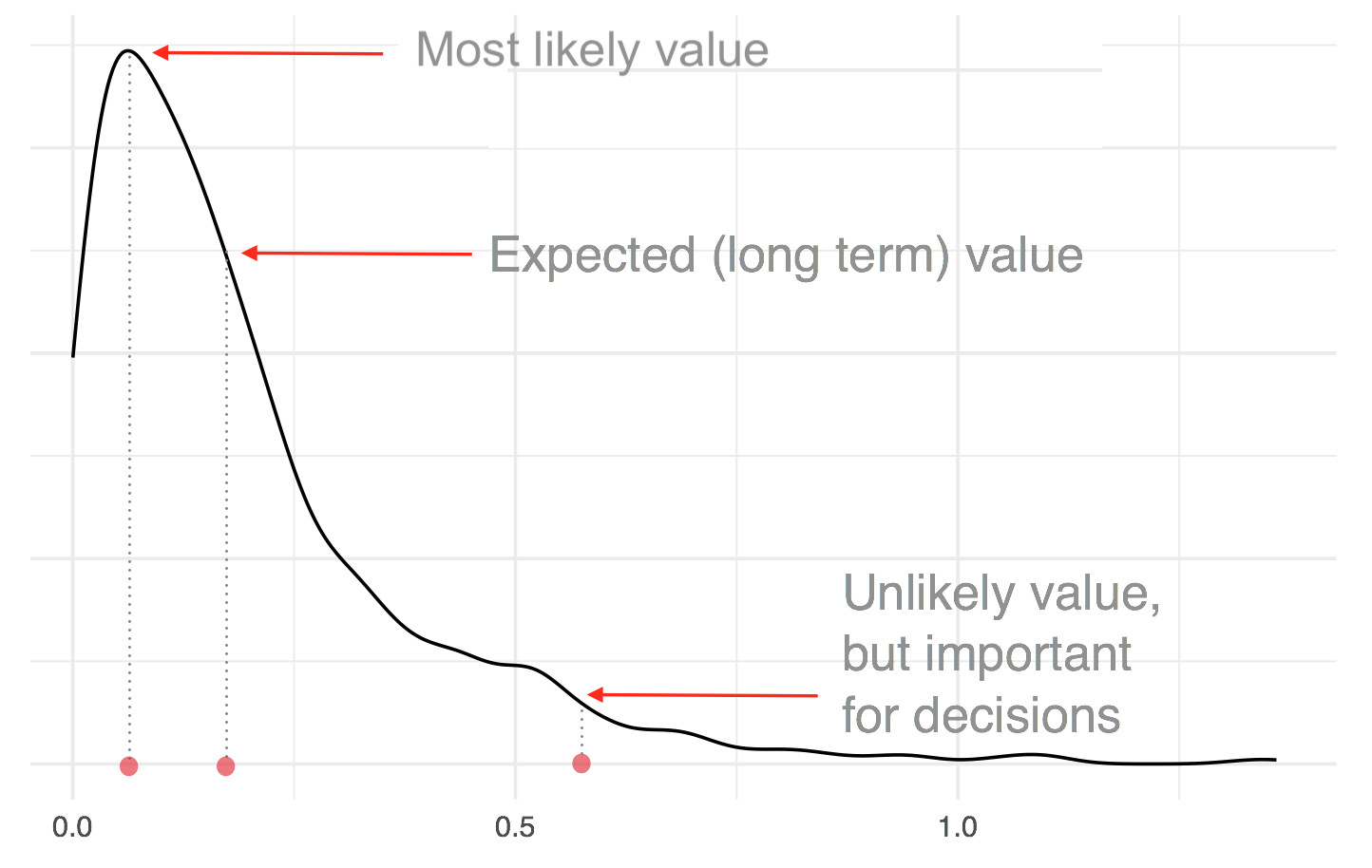
- Bayesians want to characterize the whole distribution, a much more ambitious goal
- Suppose we want to characterize the following function, which represents some distribution of the unknown parameter:
Basic Regression Setup
- There are lots of possible ways to set up a regression
- One predictor: \(y = a + bx + \epsilon\)
- Multiple predictors: \(y = b_0 + b_1x_1 + b_2x_2 + \cdots + b_nx_n + \epsilon\), which we can write as \(y = X\beta + \epsilon\)
- Models with interactions: \(y = b_0 + b_1x_1 + b_2x_2 + b_3x_1x_2 + \epsilon\)
- Non-linear models like: \(\log(y) = a + b\log(x) + \epsilon\)
- Generalized Linear Models (GLMs), which can, for example, fit binary data, categorical data, and so on
- Nonparametric models that are popular in Machine Learning that can learn flexible, functional forms between \(y\) and \(X\)
- The latter is not a panacea — if you have a good or good enough functional form, predictions will generally be better and the model will generalize better
Most of the material in this section comes from Regression and Other Stories by Gelman et al.
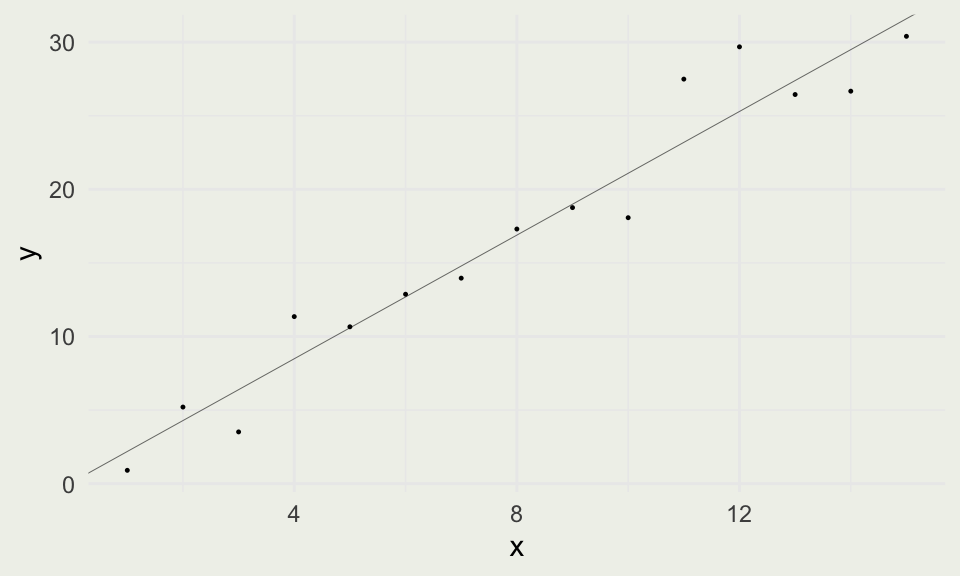
Simulating a Simple Regression
- Fitting simulated data is a good start for an analysis
Simulating a Simple Regression
- We extract our betas using the
coef()R function
KidIQ Dataset
- Data from a survey of adult American women and their children (a subsample from the National Longitudinal Survey of Youth).
Single Binary Predictor
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_hs
observations: 434
predictors: 2
------
Median MAD_SD
(Intercept) 77.6 2.0
mom_hs 11.8 2.4
Auxiliary parameter(s):
Median MAD_SD
sigma 19.9 0.7
------
* For help interpreting the printed output see ?print.stanreg
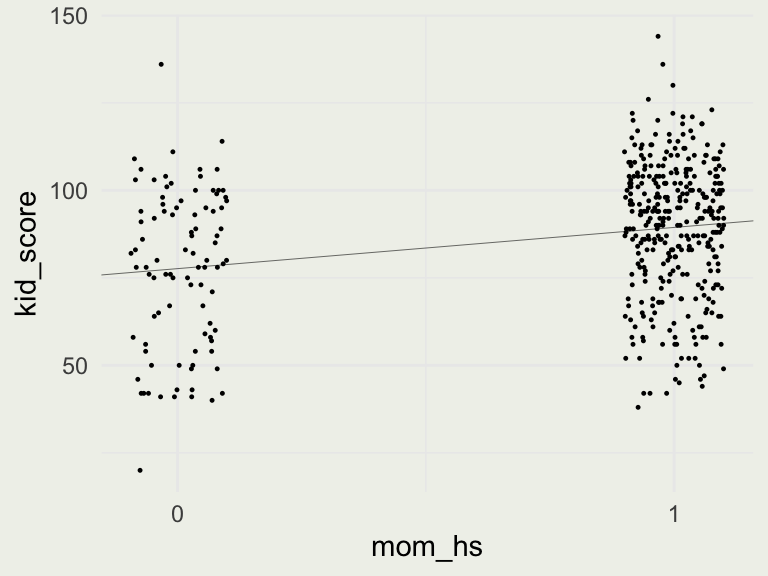
* For info on the priors used see ?prior_summary.stanreg- Our model is \(\text{kid_score} = 78 + 12 \cdot \text{mom_hs} + \epsilon\)
- What is the average IQ of kids whose mothers did not complete high school? (in this dataset)
- What is the average IQ of kids whose mothers completed high school?
- Did high school cause the IQ change?
Single Binary Predictor
Single Continous Predictor
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_iq
observations: 434
predictors: 2
------
Median MAD_SD
(Intercept) 26.0 6.1
mom_iq 0.6 0.1
Auxiliary parameter(s):
Median MAD_SD
sigma 18.3 0.6
------
* For help interpreting the printed output see ?print.stanreg
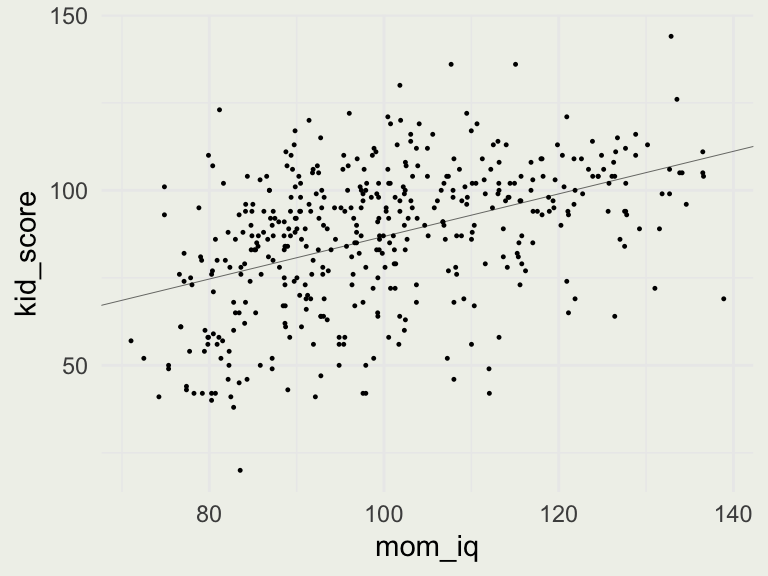
* For info on the priors used see ?prior_summary.stanreg- Our model is \(\text{kid_score} = 26 + 0.6 \cdot \text{mom_iq}+ \epsilon\)
- How much do kids’ scores improve when maternal IQ differs by 10 points?
- What is the meaning of the intercepts = 26 here?
Simple Transformations
- We will create a new variable called
mom_iq_c, which stands for centered - The transformation: \(\text{mom_iq_c}_i = \text{mom_iq}_i - \overline{\text{mom_iq}}\)
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_iq_c
observations: 434
predictors: 2
------
Median MAD_SD
(Intercept) 86.8 0.9
mom_iq_c 0.6 0.1
Auxiliary parameter(s):
Median MAD_SD
sigma 18.3 0.6
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg- Our model is \(\text{kid_score} = 87 + 0.6 \cdot \text{mom_iq_c}+ \epsilon\)
- What is the meaning of the intercepts in this model?
Single Continous Predictor
Combining the Predictors
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_hs + mom_iq_c
observations: 434
predictors: 3
------
Median MAD_SD
(Intercept) 82.2 1.9
mom_hs 6.0 2.2
mom_iq_c 0.6 0.1
Auxiliary parameter(s):
Median MAD_SD
sigma 18.2 0.6
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg- Our model is now: \(\text{kid_score} = 82 + 6 \cdot \text{mom_hs} + 0.6 \cdot \text{mom_iq_c}+ \epsilon\)
- Write down the interpretation of each coefficient
Combining the Predictors
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_hs + mom_iq
observations: 434
predictors: 3
------
Median MAD_SD
(Intercept) 25.8 5.9
mom_hs 6.0 2.3
mom_iq 0.6 0.1
Auxiliary parameter(s):
Median MAD_SD
sigma 18.2 0.6
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg- Our model is: \(\text{kid_score} = 26 + 6 \cdot \text{mom_hs} + 0.6 \cdot \text{mom_iq}+ \epsilon\)
- For moms that did not complete high school, the line \(y = 26 + 0.6 \cdot \text{mom_iq}\)
- For moms that did: \(\text{kid_score} = 26 + 6 \cdot 1 + 0.6 \cdot \text{mom_iq} = 32 + 0.6 \cdot \text{mom_iq}\)
Visualizing the Fit With Two Predictors
p <- ggplot(aes(mom_iq, kid_score), data = kidiq)
p + geom_point(aes(color = as.factor(mom_hs)), size = 0.2) +
labs(color = "Mom HS") +
geom_abline(intercept = coef(fit6)[1], slope = coef(fit6)[3], linewidth = 0.1, color = 'red') +
geom_abline(intercept = coef(fit6)[1] + coef(fit6)[2], slope = coef(fit6)[3], linewidth = 0.1, color = 'blue')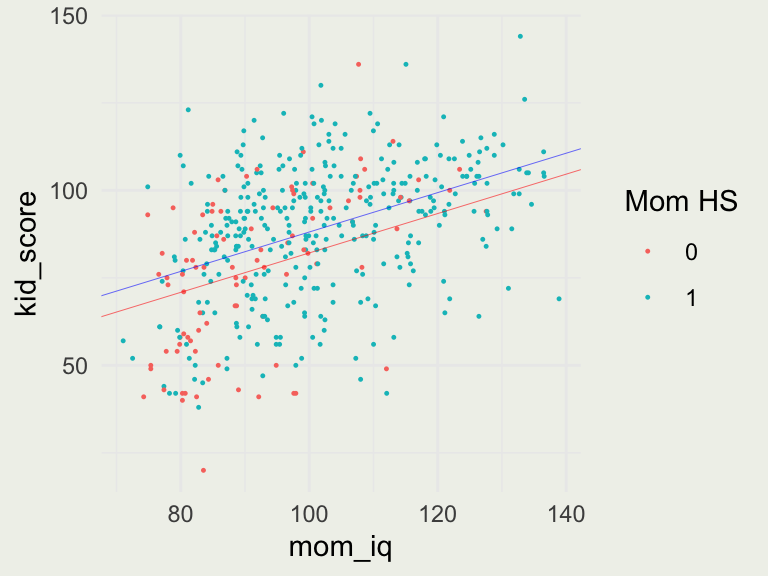
Adding Interactions
- In the previous model, we forced the slopes of mothers who completed and did not complete high school to be the same — the only difference was the intercept
- To allow the slopes to vary, we include an interaction term
stan_glm
family: gaussian [identity]
formula: kid_score ~ mom_hs + mom_iq + mom_hs:mom_iq
observations: 434
predictors: 4
------
Median MAD_SD
(Intercept) -10.6 13.7
mom_hs 50.2 15.1
mom_iq 1.0 0.1
mom_hs:mom_iq -0.5 0.2
Auxiliary parameter(s):
Median MAD_SD
sigma 18.0 0.6
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg- Our model is: \(\text{kid_score} = -10 + 49 \cdot \text{mom_hs} + 1 \cdot \text{mom_iq} - 0.5 \cdot \text{mom_hs} \cdot \text{mom_iq} + \epsilon\)
- To figure out the slopes, we consider two cases
- For \(\text{mom_hs} = 0\), the line is: \(\text{kid_score} = -10 + 1 \cdot \text{mom_iq}\)
- For \(\text{mom_hs} = 1\), the line is: \(\text{kid_score} = -10 + 49 \cdot 1 + 1 \cdot \text{mom_iq} - 0.5 \cdot 1 \cdot \text{mom_iq} = 39 + 0.5\cdot \text{mom_iq}\)
Visualizing Interactions
p <- ggplot(aes(mom_iq, kid_score), data = kidiq)
p + geom_point(aes(color = as.factor(mom_hs)), size = 0.2) +
labs(color = "Mom HS") +
geom_abline(intercept = coef(fit7)[1], slope = coef(fit7)[3], linewidth = 0.1, color = 'red') +
geom_abline(intercept = coef(fit7)[1] + coef(fit7)[2], slope = coef(fit7)[3] + coef(fit7)[4], linewidth = 0.1, color = 'blue')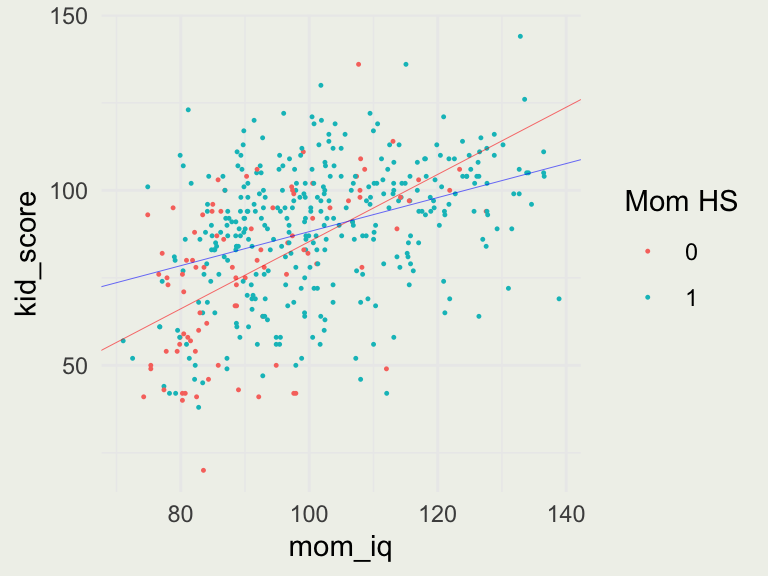
Interpreting Coefficients in Complex Non-Linear Models
- Models that have many parameters, many interactions, and are non-linear are difficult to interpret by looking at parameters marginally (i.e., one at a time)
- For this reason, we typically rely on assessing how changes to model inputs affect model predictions, not the impact of each parameter
- Predictions take all the parameter values and their interactions into account
Uncertainty and Prediction
- We return to our simple linear regression model: \(\text{kid_score} = 26 + 0.6 \cdot \text{mom_iq}+ \epsilon\)
- We can extract the simulations of all plausible parameter values (called a posterior distribution in Bayesian analysis)
Uncertainty and Prediction
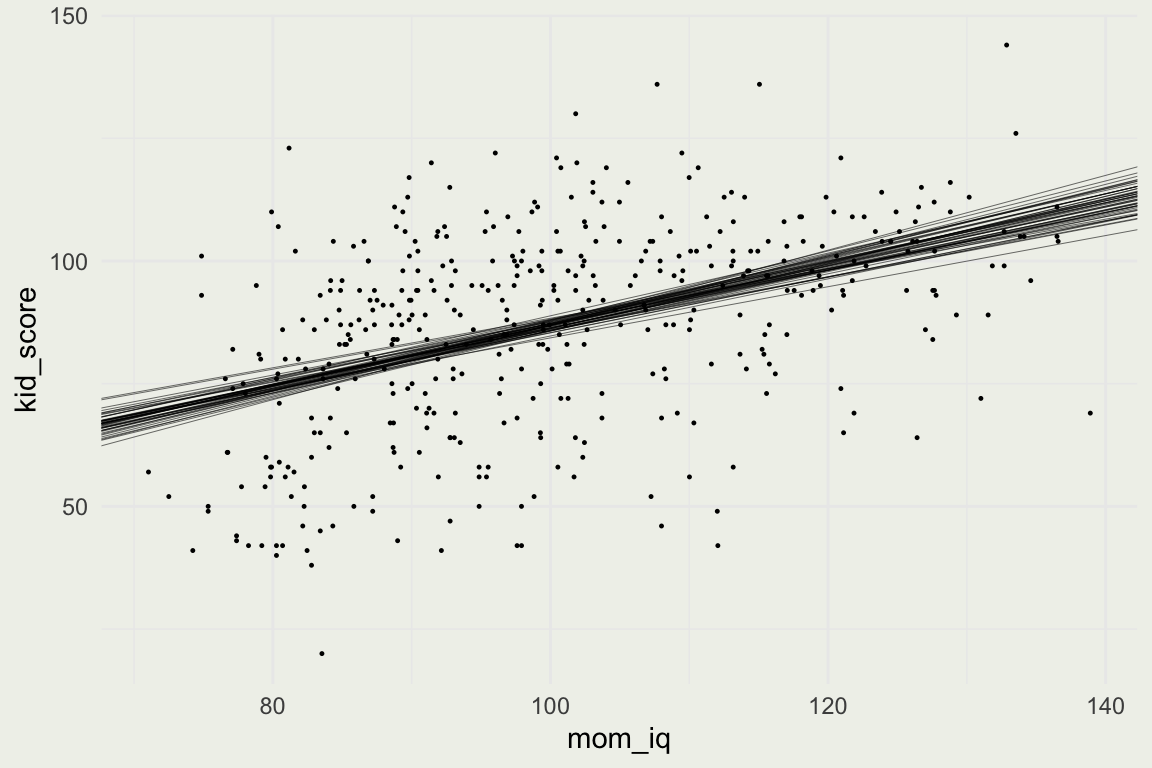
- Using these simulations we display 50 plausible regression lines
Uncertainty and Prediction
- We can compute inferential uncertainty in all the parameters directly
- We can also compute event probabilities
(Intercept) mom_iq sigma
26.0236767 0.6078977 18.2714868 (Intercept) mom_iq sigma
6.10031655 0.06062729 0.61543505 parameters
(Intercept) mom_iq sigma
25% 21.75756 0.5681001 17.86710
75% 30.06156 0.6504483 18.69458 parameters
(Intercept) mom_iq sigma
5% 15.72707 0.5106528 17.32535
95% 35.77134 0.7095889 19.33193[1] 0.55Types of Prediction
- Point prediction: \(\hat{a} + \hat{b}x^{\text{new}}\) — predicting expected average \(y\) — don’t recommend
- Linear predictor with uncertainty in \(a\) and \(b\): \(a + bx^{\text{new}}\) — predicting uncertainty around the expexted average value of \(y\) — typically used to assess treatment effect
- Predictive distribution: \(a + bx^{\text{new}} + \epsilon\) — predictive uncertainty around a new \(y\) — used to predict for a new individual (not ATT)
Prediction in RStanArm
- We observe a new kid whose mom’s IQ = 120
- Point prediction is shown in Red, 90% uncertainty around the average kid score in Blue, and 90% predictive uncertainty for a new kid is in Yellow.
1
98.9816 5% 95%
96.5370 101.4359 5% 95%
68.88551 128.75195 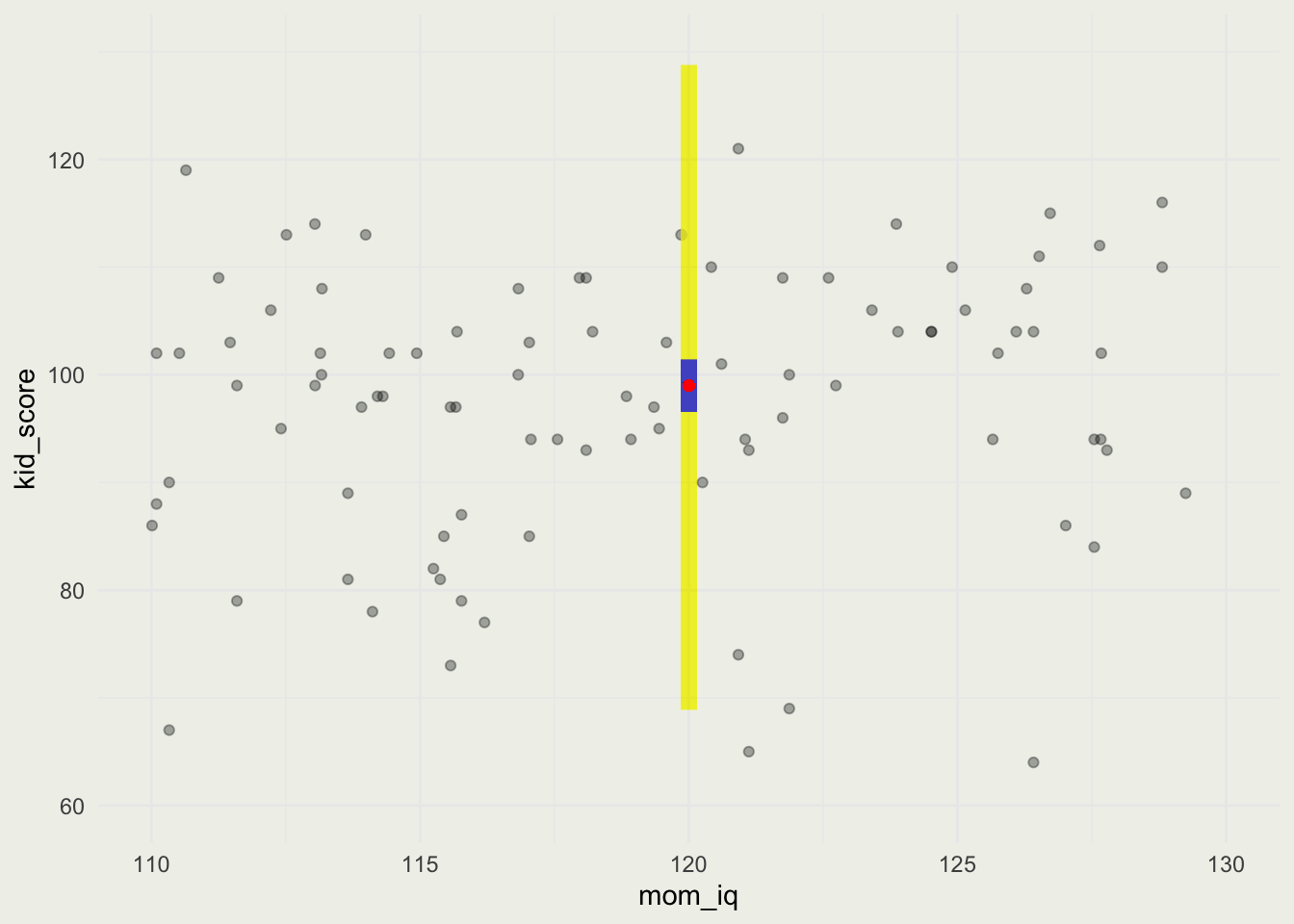
Linear Regression Assumptions
Presented in the order of importance
- Validity of the model for the question at hand
- Representativeness and valid scope of inference
- Additivity and linearity
- Independence of errors
- Equal variance of errors
- Normality of errors
10+ quick tips to improve your regression modeling
This advice comes from “Regression and Other Stories” by Gelman and Hill
- Think generatively
- Think about what you are measuring
- Think about variation, replication
- Forget about statistical significance
- Graph the relevant and not the irrelevant
- Interpret regression coefficients as comparisons
- Understand statistical methods using fake-data simulation
- Fit many models
- Set up a computational workflow
- Use transformations
- Do causal inference in a targeted way, not as a byproduct of a large regression
- Learn methods through live examples
Thanks for being part of my class!

- Lectures: https://ericnovik.github.io/smac.html
- Keep in touch:
- eric.novik@nyu.edu
- https://www.linkedin.com/in/enovik/
- Twitter: @ericnovik
- Personal blog: https://ericnovik.com/